library(tidyverse)
# Just use the first ten rows of mpg
mpg_small <- mpg |>
slice(1:10)
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point()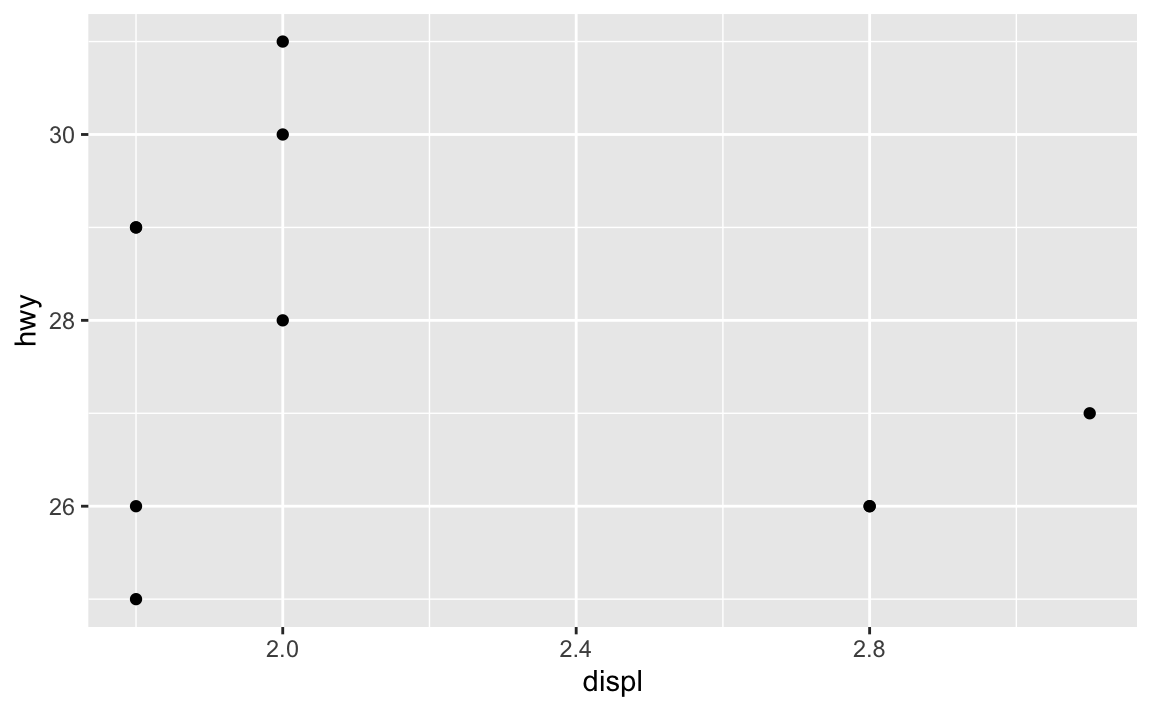
Reminder! The final deadline for all assignments other than the final project is Tuesday, July 29 at 11:59 PM
Tuesday July 15, 2025 at 9:44 AM
geom_text() and annotate(geom = "text")?The short version: geom_text() puts text on every point in the dataset; annotate() lets you add one item of text somewhere in the plot.
All the different geom_*() functions show columns from a dataset mapped onto some aesthetic. You have to have set aesthetics with aes() to use them, and you’ve been using things like geom_point() all semester:
library(tidyverse)
# Just use the first ten rows of mpg
mpg_small <- mpg |>
slice(1:10)
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point()
If we add geom_text(), we’ll get a label at every one of these points. Essentially, geom_text() is just like geom_point(), but instead of adding a point, it adds text.
Here we’ve mapped the model column from the dataset to the label aesthetic, so it’s showing the name of the model. You can map any other column too, like label = hwy:
Where this trips people up is when you want to add annotations. You might say “Hey! I want to add an label that says ‘These are cars!’”, so you do this:
Oops. That’s no what you want! You’re getting the “These are cars!” annotation, but it’s showing up at every single point. That’s because you’re using geom_text()—it adds text for every row in the dataset.
In this case, you want to use annotate() instead. As you learned in the lesson, annotate() lets you add one single geom to the plot without needing any data mapped to it. You supply the data yourself.
For example, there’s an empty space in this plot in the upper right corner. Let’s put a label at displ = 2.8 and hwy = 30, since there’s no data there:
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point() +
annotate(geom = "text", x = 2.8, y = 30, label = "These are cars!")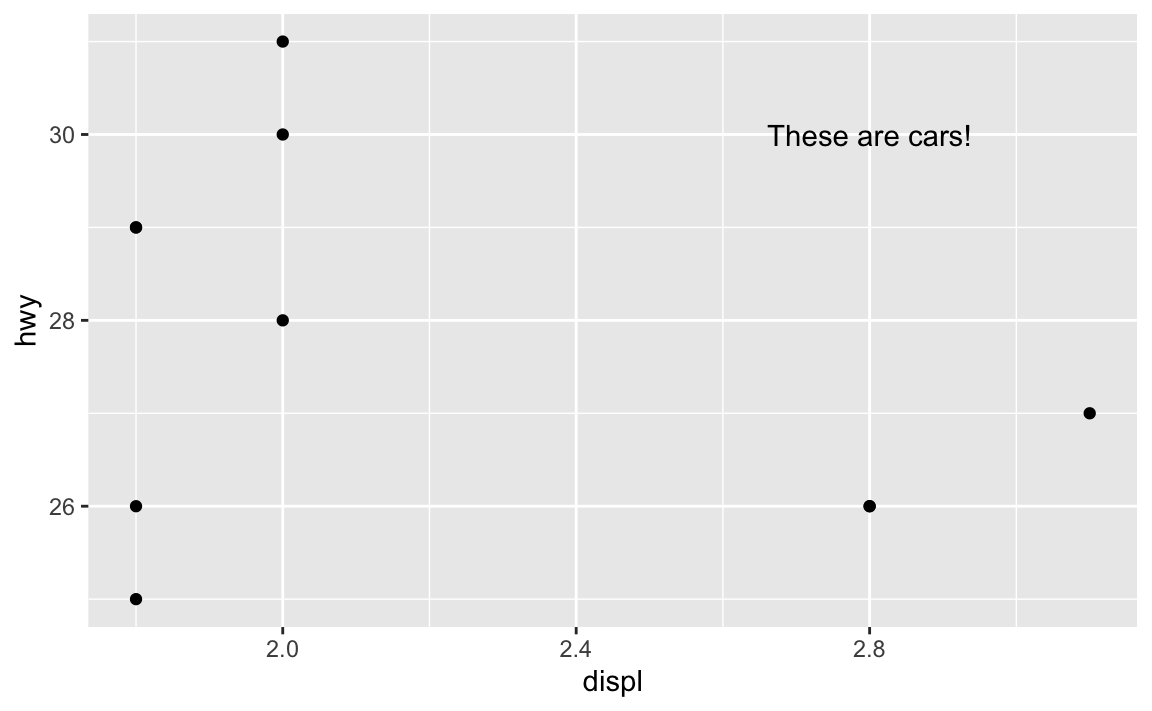
Now there’s just one label at the exact location we told it to use.
With annotate(), you have to specify all your own aesthetics manually, like x and y and label in the example above.How did we know that the text geom needed those?
The help page for every geom has a list of possible and required aesthetics. Aesthetics that are in bold are required. Here’s the list for geom_text()—it needs an x, y, and label, and it can do a ton of other things like color, alpha, size, fontface, and so on:
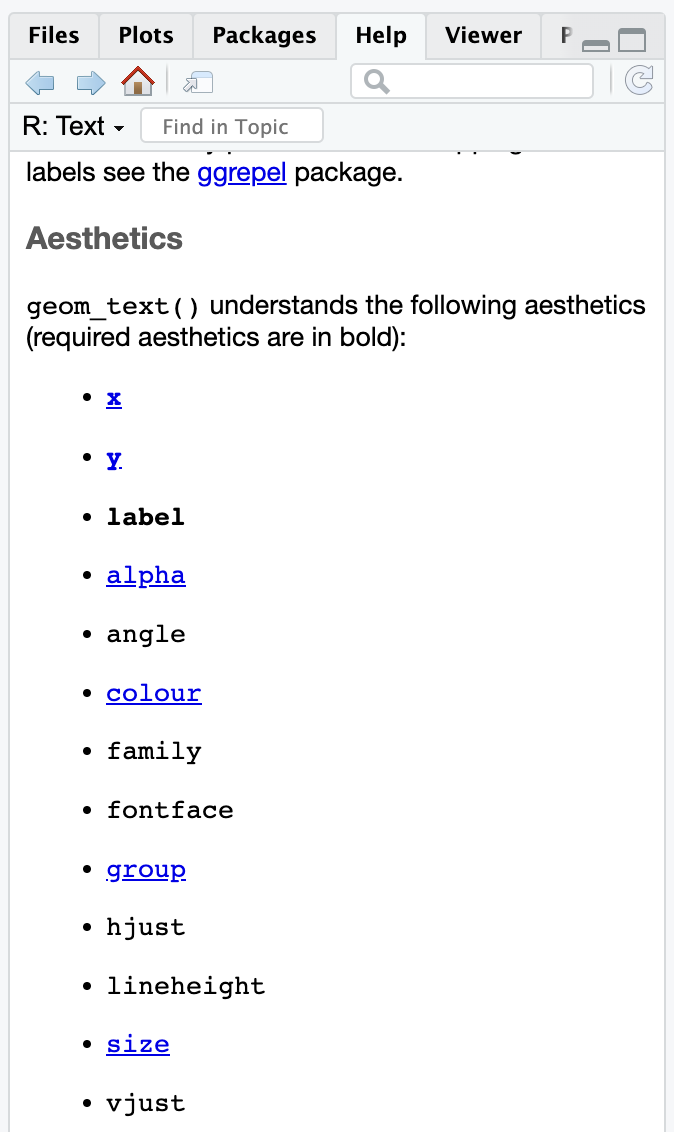
geom_textIf you want to stick a rectangle on a plot with annotate(geom = "rect"), you need to look at the help page for geom_rect() to see how it works and which aesthetics it needs.
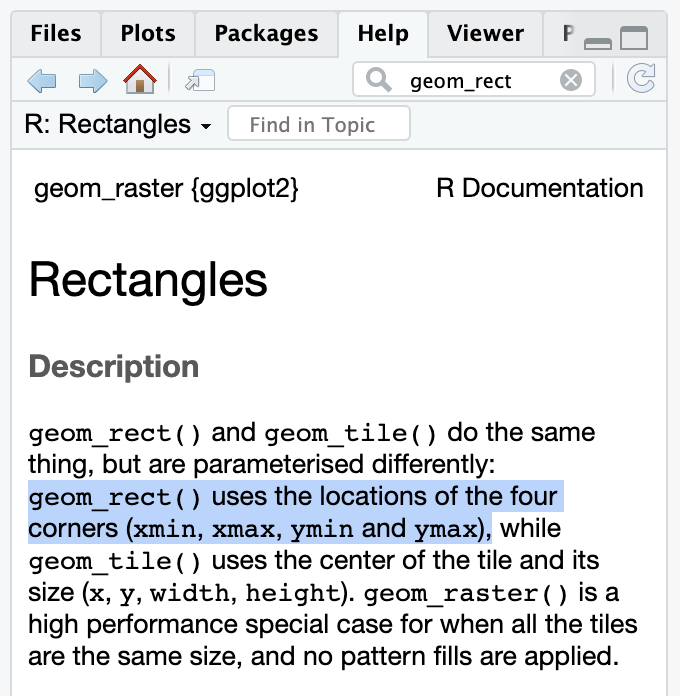
geom_rectgeom_rect() needs xmin, xmax, ymin, and ymax, and it can also do alpha, color (for the border), fill (for the fill), linewidth, and some other things:
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point() +
annotate(
geom = "rect",
xmin = 2.5, xmax = 3.1, ymin = 29, ymax = 31,
fill = "red", alpha = 0.2
) +
annotate(geom = "text", x = 2.8, y = 30, label = "These are cars!")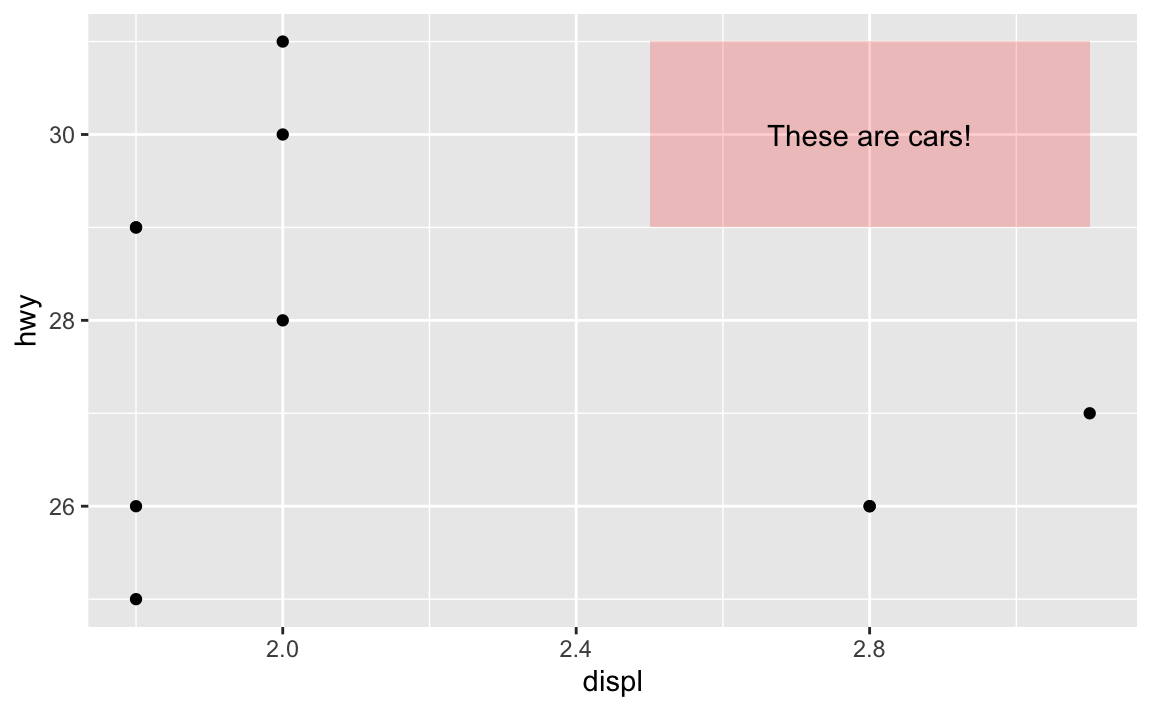
In that same help page, it mentions that geom_tile() is an alternative to geom_rect(), where you define the center of the rectangle with x and y and define the width and the height around that center. This is helpful for getting rectangles centered around certain points. I just eyeballed the 2.5, 3.1, 29, and 31 up there ↑ to get the rectangle centered behind the text. I can get it precise with geom_tile() instead:
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point() +
annotate(
geom = "tile",
x = 2.8, y = 30, width = 0.5, height = 2,
fill = "red", alpha = 0.2
) +
annotate(geom = "text", x = 2.8, y = 30, label = "These are cars!")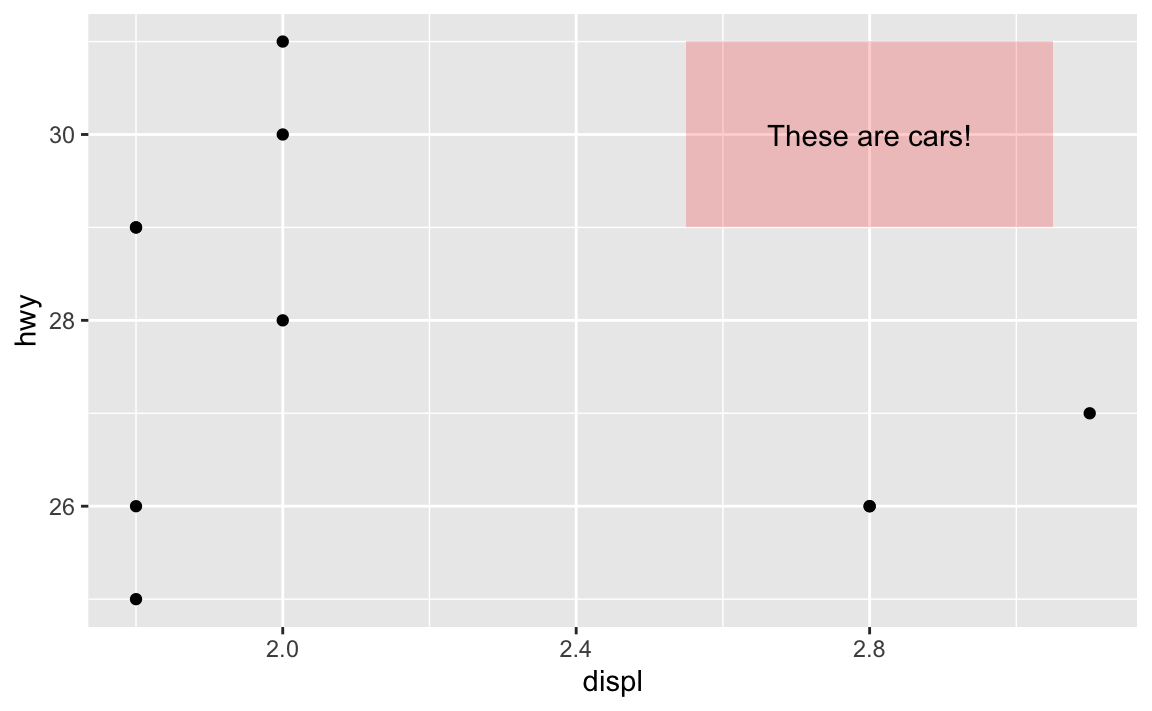
So always check the “Aesthetics” section of a geom’s help page to see what you can do with it!
Yeah, this is super annoying. Like, what if you want a label to be horizontally centered in the plot, and have it near the top, maybe like 80% of the way to the top? Here’s how to eyeball it:
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point() +
annotate(geom = "text", x = 2.4, y = 30, label = "Middle-ish label")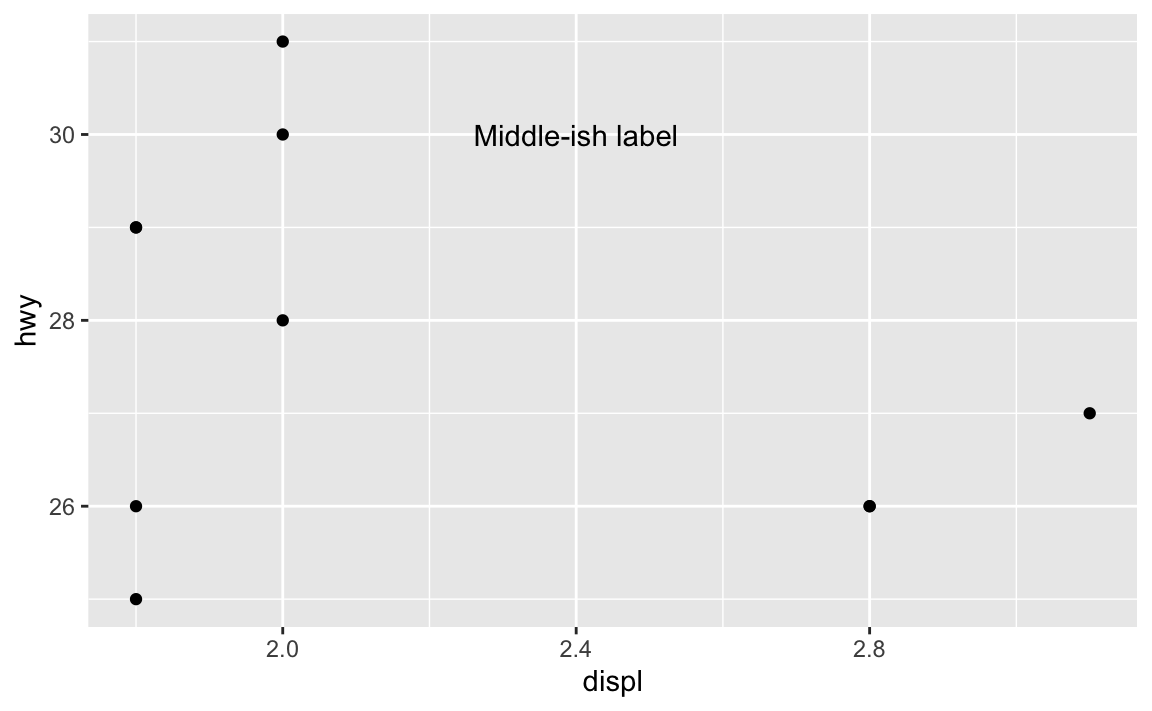
But that’s not actually centered—the x would need to be something like 2.46 or something ridiculous. And this isn’t very flexible. If new points get added or the boundareies of the axes change, that label will most definitely not be in the middle:
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point() +
annotate(geom = "text", x = 2.4, y = 30, label = "Middle-ish label") +
coord_cartesian(xlim = c(1, 3), ylim = c(25, 35))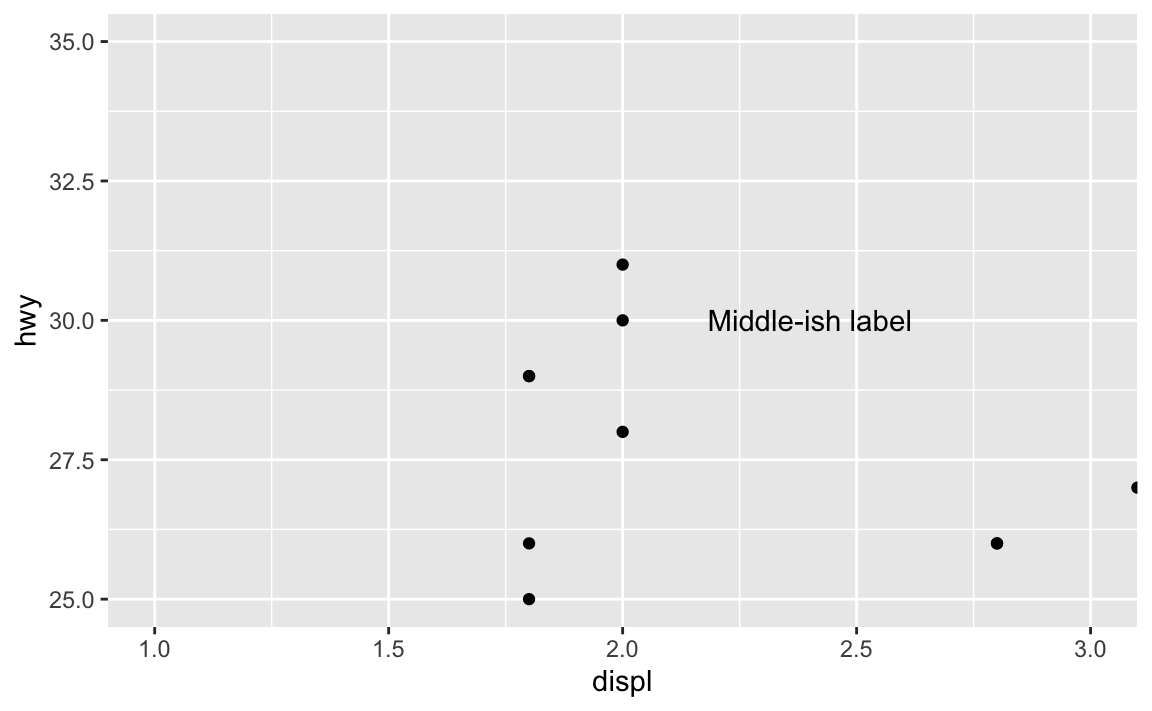
Fortunately there’s a way to ignore the values in the x and y axes and instead use relative positioning (see this for more). If you use a special I() function, you can define positions by percentages, so that x = I(0.5) will put the annotation at the 50% position in the plot, or right in the middel. y = I(0.8) will put the annotation 80% of the way up the y-axis:
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point() +
annotate(geom = "text", x = I(0.5), y = I(0.8), label = "Exact middle label")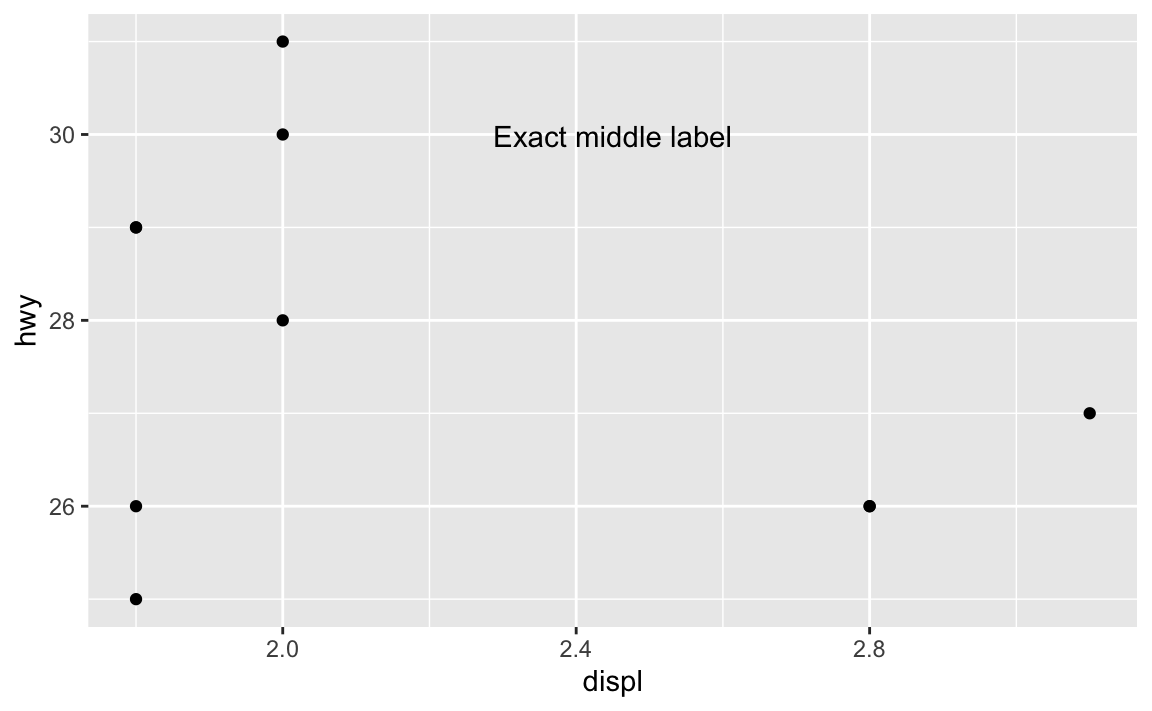
Now that’s exactly centered and 80% up, and will be regardless of how it’s zoomed. If we adjust the axes, it’ll still be there:
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point() +
annotate(geom = "text", x = I(0.5), y = I(0.8), label = "Exact middle label") +
coord_cartesian(xlim = c(1, 3), ylim = c(25, 35))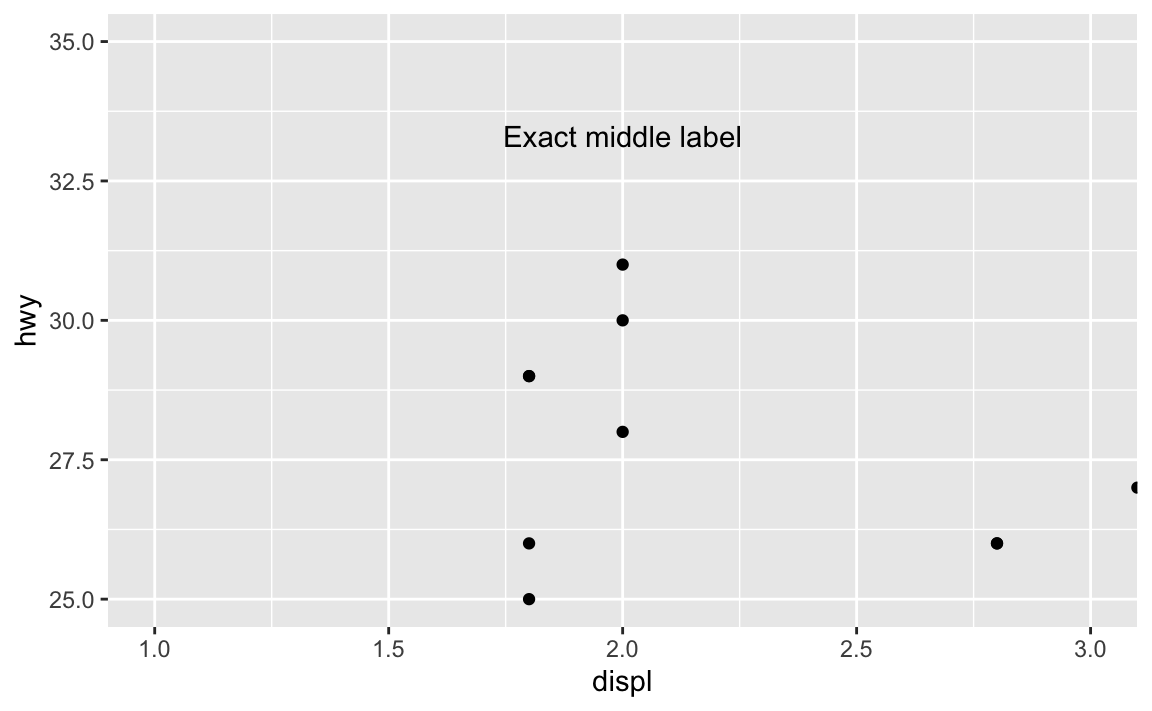
Want a rectangle to go all around the plot with corners at 10% and 90%, with a label that’s centered and positioned at 90% so it looks like it’s connected to the rectangle? Easy!
geom_text() and geom_label()?They’re the same, except geom_label() adds a border and background to the text. See this from the help page for geom_label():
geom_text()adds only text to the plot.geom_label()draws a rectangle behind the text, making it easier to read.
Which is better to use? That’s entirely context-dependent—there are no right answers.
update_geom_defaults() do?In the FAQs for sessions 5 and 6, I mentioned the theme_set() function, which lets you set a theme for all plots in a document:
# Make all plots use theme_minimal()
theme_set(theme_minimal())
# This now uses theme_bw without needing to specify it
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point() +
geom_smooth(method = "lm", se = FALSE)
Let’s say I want those points to be a little bigger and be squares, and I want the fitted line to be thicker. I can change those settings in the different geom layers:
ggplot(mpg_small, aes(x = displ, y = hwy)) +
geom_point(size = 3, shape = "square") +
geom_smooth(method = "lm", se = FALSE, linewidth = 2)
What if I want all the points and lines in the document to be like that: big squares and thick lines? I’d need to remember to add those settings every time I use geom_point() or geom_smooth().
Or, even better, I can change all the default geom settings once:
Now every instance of geom_point() and geom_smooth() will use those settings:
In the example for session 9, I used update_geom_defaults() to change the font for all the text and label geoms. Without that, I’d need to include family = "IBM Plex Sans" in every single layer that used text or labels.
Oh man, this is a tricky one!
Here’s a little plot of penguin stuff faceted by penguin species:
library(palmerpenguins)
penguins <- palmerpenguins::penguins |> drop_na(sex)
ggplot(penguins, aes(x = bill_length_mm, y = body_mass_g, color = species)) +
geom_point() +
guides(color = "none") + # No need for a legend since we have facets
facet_wrap(vars(species))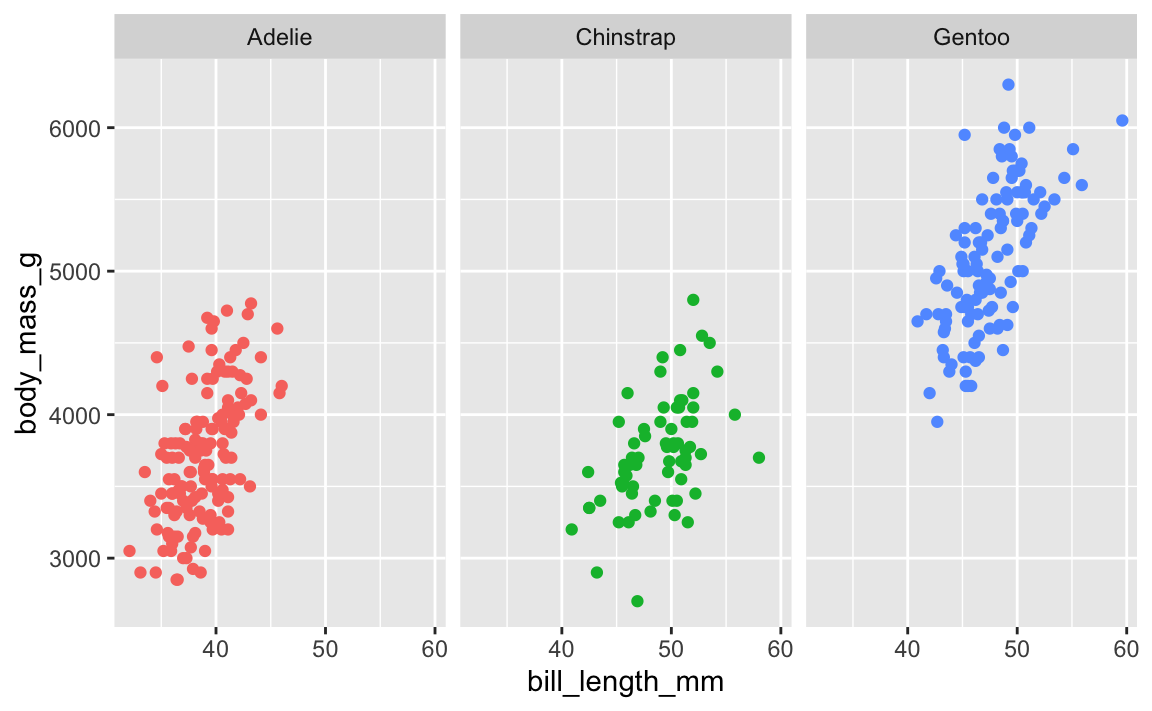
Cool. Now let’s add a label pointing out that Gentoos are bigger than the others:
ggplot(penguins, aes(x = bill_length_mm, y = body_mass_g, color = species)) +
geom_point() +
guides(color = "none") + # No need for a legend since we have facets
facet_wrap(vars(species)) +
annotate(geom = "label", x = 45, y = 3500, label = "big penguins!")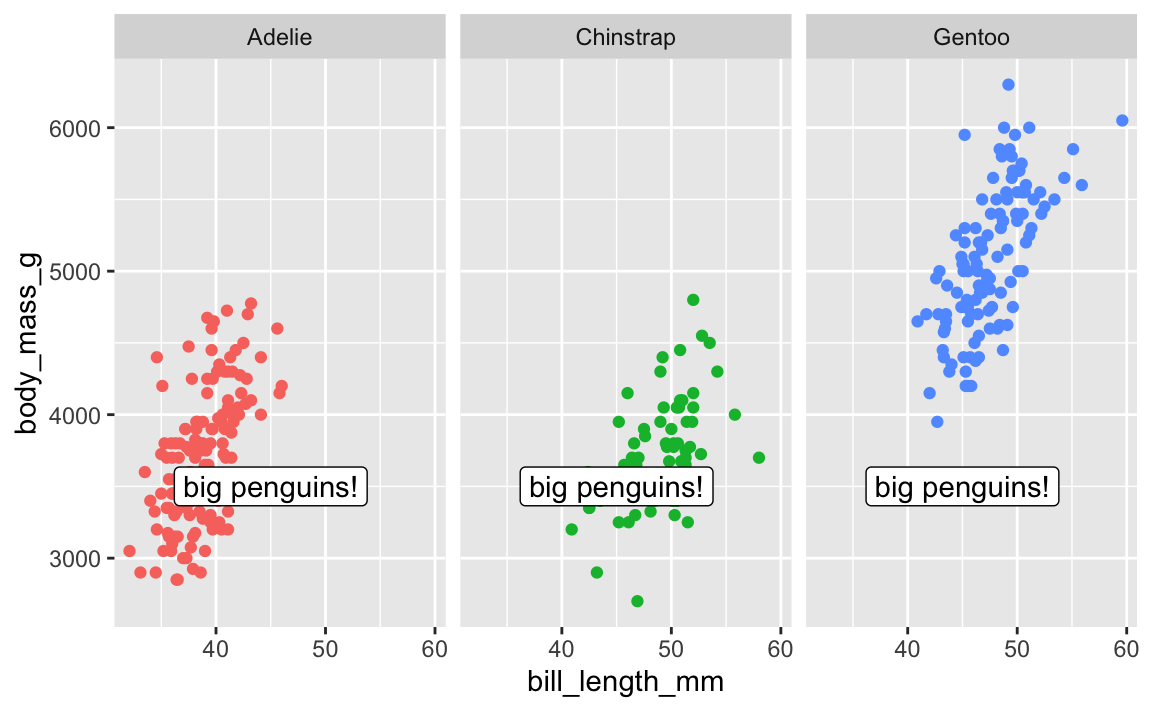
Oops. That label appears in every facet! There is no built-in way to specify that that annotate() should only appear in one of the facets.
There are two solutions: one is super easy and one is more complex but very flexible.
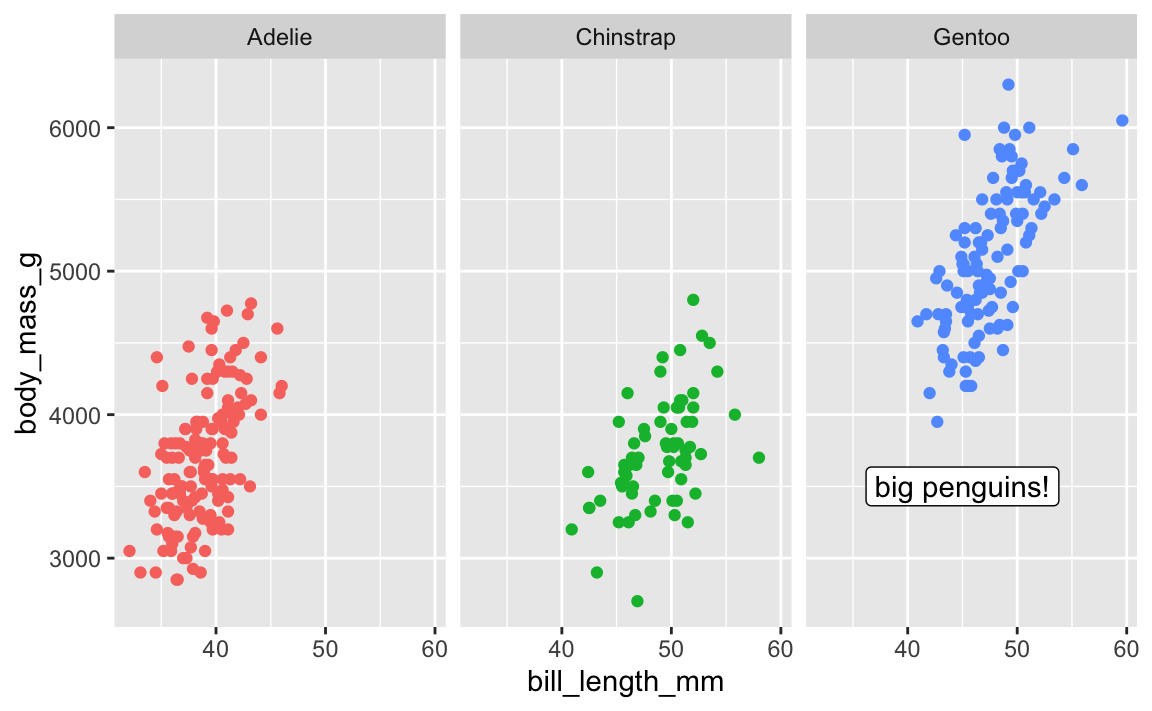
First, the easy one. There’s a package named {ggh4x} that has a bunch of really neat ggplot enhancements (like, check out nested facets! I love these and use them all the time). One function it includes is at_panel(), which lets you constrain annotate() layers to specific panels
library(ggh4x)
ggplot(penguins, aes(x = bill_length_mm, y = body_mass_g, color = species)) +
geom_point() +
guides(color = "none") + # No need for a legend since we have facets
facet_wrap(vars(species)) +
at_panel(
annotate(geom = "label", x = 45, y = 3500, label = "big penguins!"),
species == "Gentoo"
)
Next, the more complex one. With this approach, we don’t use annotate() and use geom_label() instead. I KNOW THIS FEELS WRONG—there was a whole question above about the difference between the two and I said that geom_text() is for labeling all the points in the dataset while annotate() is for adding one label.
So the trick here is that we make a tiny little dataset with the annotation details we want:
We can then plot it with geom_label(), which lets us limit the point to just the Gentoo panel:
So far this semester, most of your plots have involved one or two geom_* layers. At one point in some video (I think), I mentioned that layer order doesn’t matter with ggplot. These two chunks of code create identical plots:
ggplot(...) +
geom_point(...) +
theme_minimal(...) +
scale_fill_viridis_c(...) +
facet_wrap(...) +
labs(...)
ggplot(...) +
geom_point(...) +
labs(...) +
theme_minimal(...) +
facet_wrap(...) +
scale_fill_viridis_c(...)All those functions can happen in whatever order you want, with one exception. The order of the geom layers matters. The first geom layer you specify will be plotted first, the second will go on top of it, and so on.
Let’s say you want to have a violin plot with jittered points on top. If you put geom_point() first, the points will be hidden by the violins:
library(palmerpenguins)
penguins <- palmerpenguins::penguins |> drop_na(sex)
ggplot(penguins, aes(x = species, y = body_mass_g)) +
geom_point(position = position_jitter(seed = 1234), size = 0.5) +
geom_violin(aes(fill = species))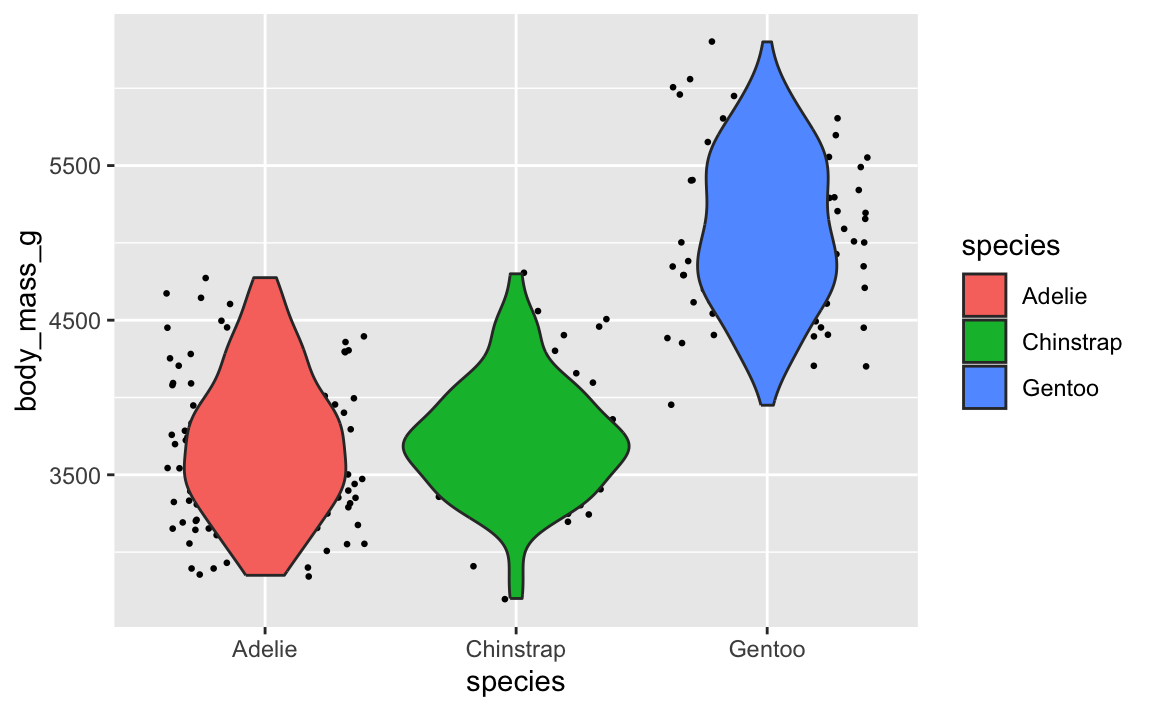
To fix it, make sure geom_violin() comes first:
ggplot(penguins, aes(x = species, y = body_mass_g)) +
geom_violin(aes(fill = species)) +
geom_point(position = position_jitter(seed = 1234), size = 0.5)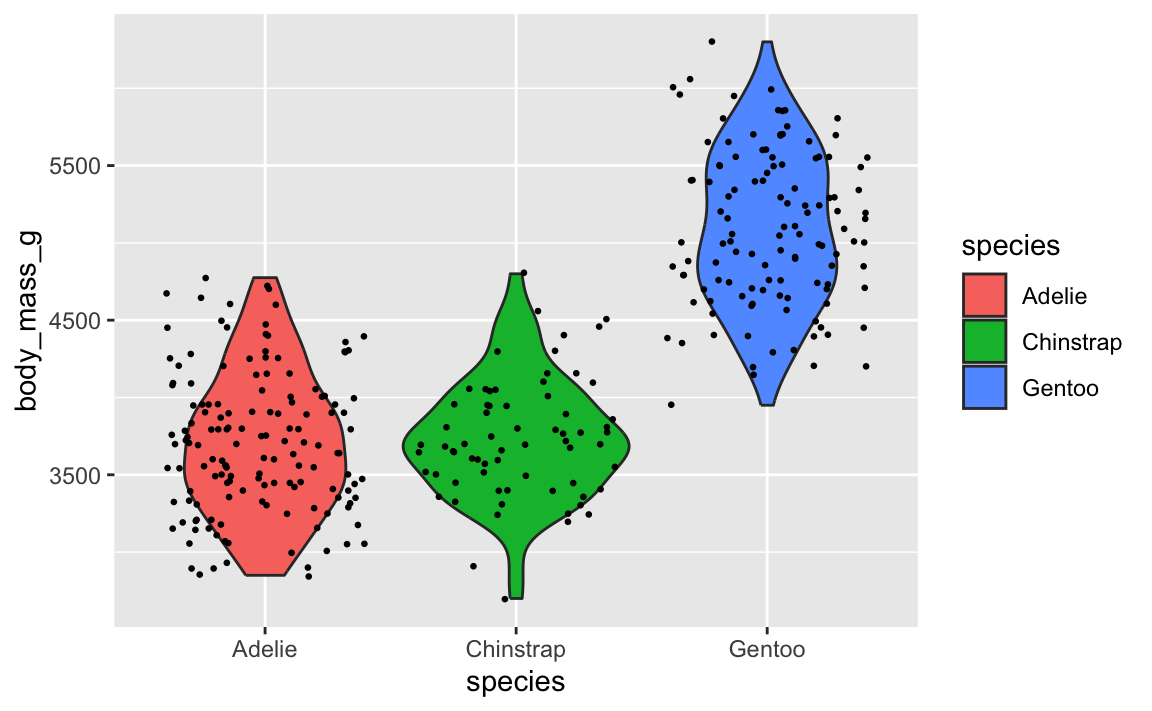
This layer order applies to annotation layers too. If you want to highlight an area of the plot, adding a rectangle after the geom layers will cover things up, like this ugly yellow rectangle here:
ggplot(penguins, aes(x = bill_length_mm, y = body_mass_g, color = species)) +
geom_point() +
annotate(geom = "rect", xmin = 40, xmax = 60, ymin = 5000, ymax = 6100,
fill = "yellow", alpha = 0.75)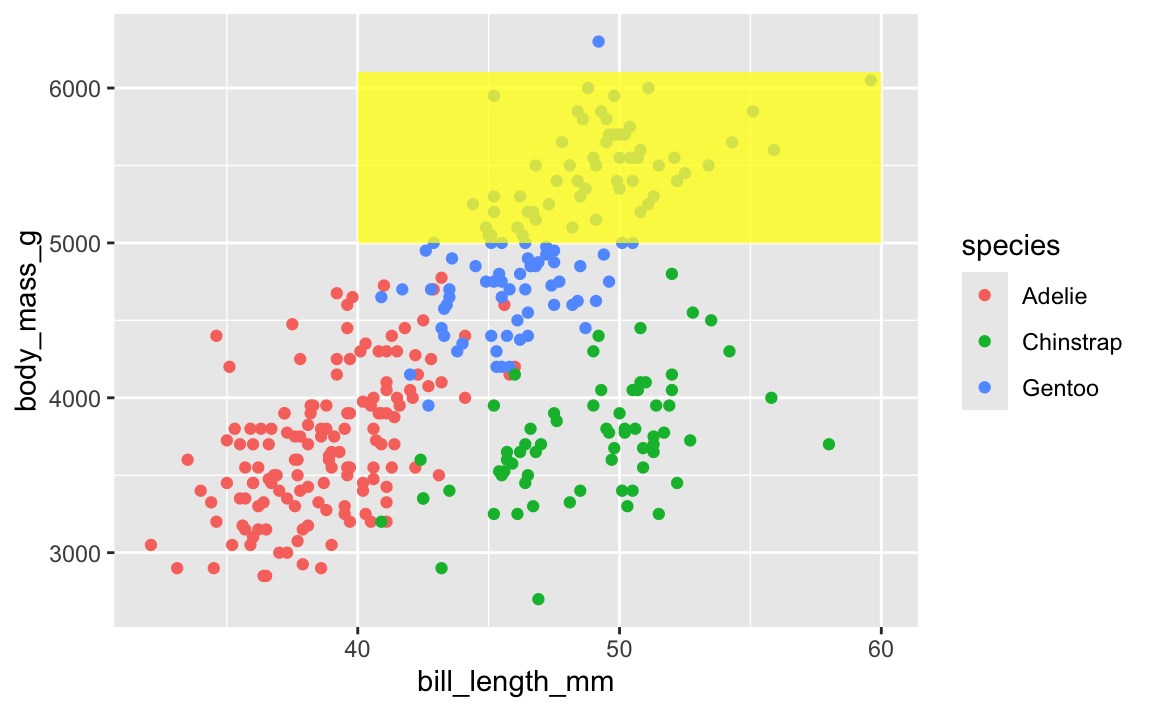
To fix that, put that annotate() layer first, then add other geoms on top:
ggplot(penguins, aes(x = bill_length_mm, y = body_mass_g, color = species)) +
annotate(geom = "rect", xmin = 40, xmax = 60, ymin = 5000, ymax = 6100,
fill = "yellow", alpha = 0.75) +
geom_point()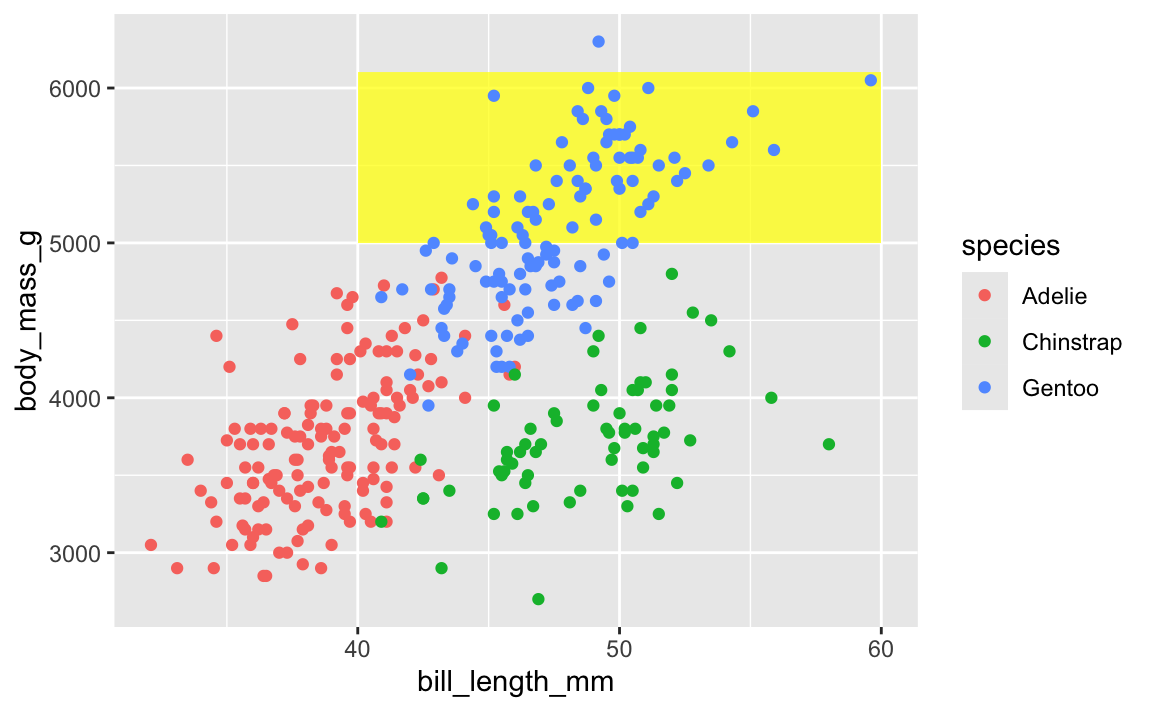
This doesn’t mean all annotate() layers should come first—if you want an extra label on top of a geom, make sure it comes after:
ggplot(penguins, aes(x = bill_length_mm, y = body_mass_g, color = species)) +
# Yellow rectangle behind everything
annotate(geom = "rect", xmin = 40, xmax = 60, ymin = 5000, ymax = 6100,
fill = "yellow", alpha = 0.75) +
# Points
geom_point() +
# Label on top of the points and the rectangle
annotate(geom = "label", x = 50, y = 5500, label = "chonky birds")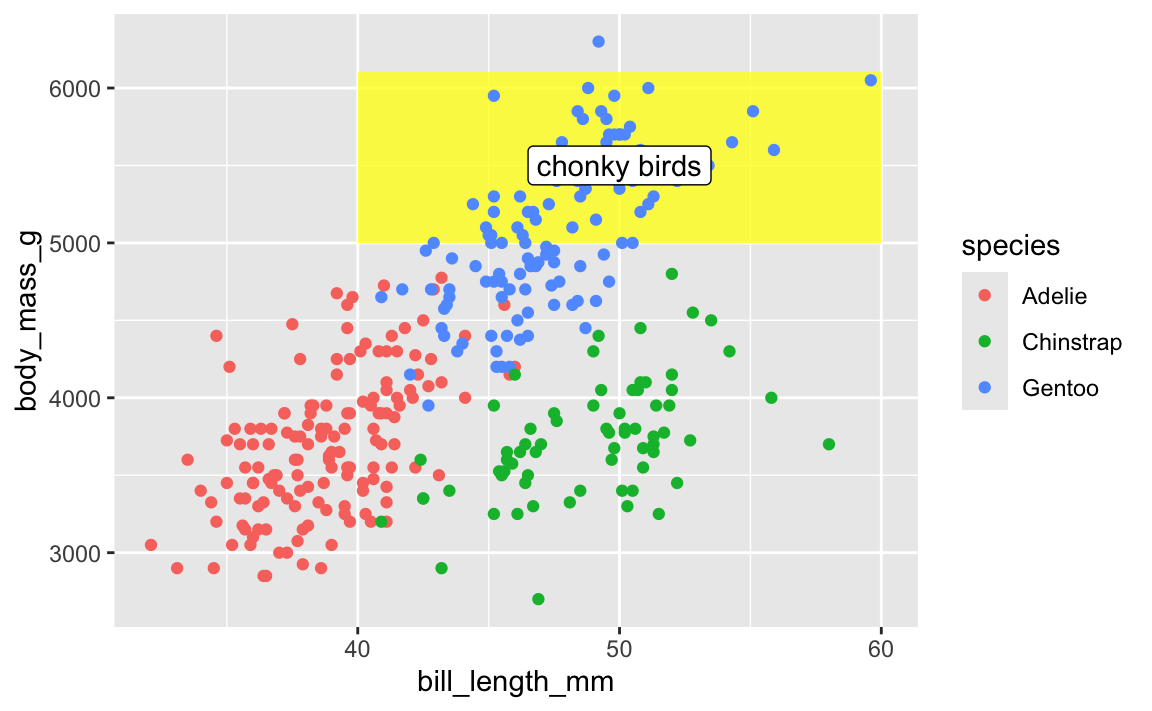
When I make my plots, I try to keep my layers in logical groups. I’ll do my geoms and annotations first, then scale adjustments, then guide adjustments, then labels, then facets (if any), and end with theme adjustments, like this:
library(scales)
ggplot(penguins, aes(x = bill_length_mm, y = body_mass_g, color = species)) +
# Annotations and geoms
annotate(geom = "rect", xmin = 40, xmax = 60, ymin = 5000, ymax = 6100,
fill = "yellow", alpha = 0.75) +
geom_point() +
annotate(geom = "label", x = 50, y = 5500, label = "chonky birds") +
# Scale adjustments
scale_x_continuous(labels = label_comma(scale_cut = cut_si("mm"))) +
scale_y_continuous(labels = label_comma(scale_cut = cut_si("g"))) +
scale_color_viridis_d(option = "plasma", end = 0.6) +
# Guide adjustments
guides(color = guide_legend(title.position = "left")) +
# Labels
labs(x = "Bill length", y = "Body mass", color = "Species:",
title = "Some title", subtitle = "Penguins!", caption = "Blah") +
# Facets
facet_wrap(vars(sex)) +
# Theme stuff
theme_minimal() +
theme(plot.title = element_text(face = "bold", size = rel(1.4)),
plot.caption = element_text(color = "grey50", hjust = 0),
axis.title.x = element_text(hjust = 0),
axis.title.y = element_text(hjust = 1),
strip.text = element_text(hjust = 0, face = "bold"),
legend.position = "bottom",
legend.justification = c(-0.04, 0),
legend.title = element_text(size = rel(0.9)))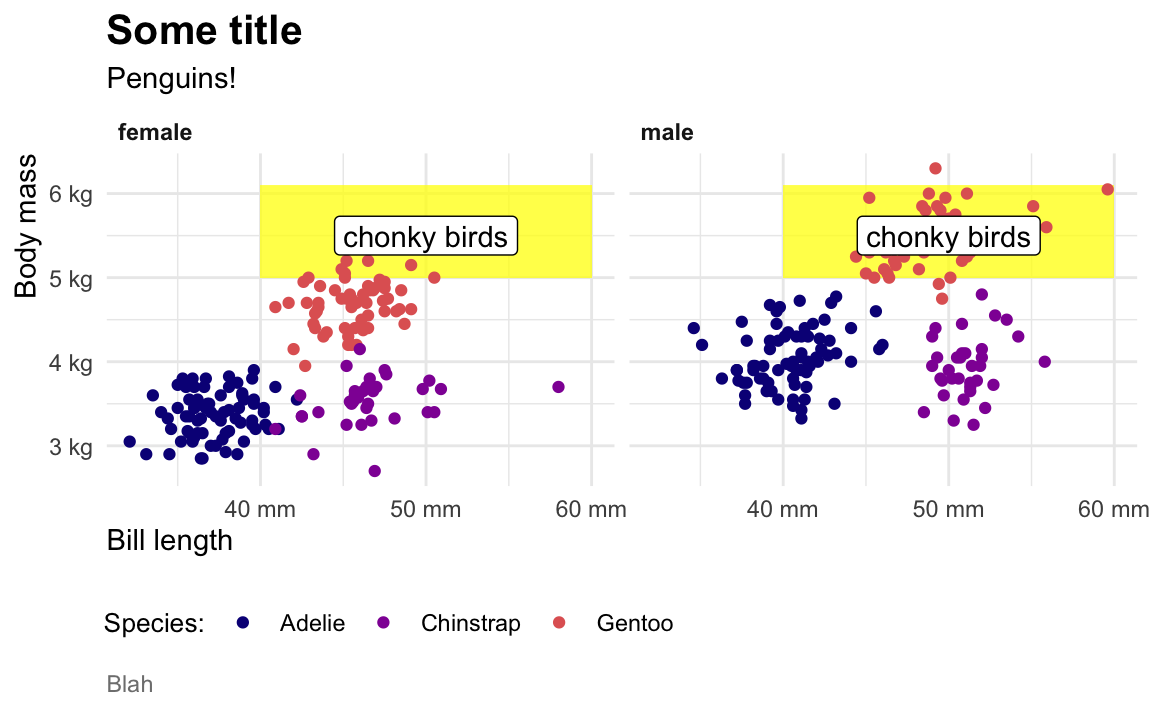
This is totally arbitrary though! All that really matters is that the geoms and annotations are in the right order and that any theme adjustments you make with theme() come after a more general theme like theme_grey() or theme_minimal(), etc.. I’d recommend you figure out your own preferred style and try to stay consistent—it’ll make your life easier and more predictable.
Yeah! A bunch of you ran into this! Here’s a little illustration using the unemployment data from exercise 8. We’ll rank all the states in the west by their unemployment rates in 2006 and 2009 and then see which ones had the biggest jumps in ranking.
Before doing any pivoting, let’s look at the data really quick:
unemployment_west_rankings <- unemployment |>
filter(region == "West", year %in% c(2006, 2009), month_name == "January") |>
group_by(year) |>
mutate(ranking = rank(unemployment)) |>
ungroup() |>
select(state, region, division, year, unemployment, ranking)
unemployment_west_rankings
## # A tibble: 26 × 6
## state region division year unemployment ranking
## <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 Alaska West Pacific 2009 7.1 7.5
## 2 Alaska West Pacific 2006 6.9 13
## 3 Arizona West Mountain 2009 8.4 10
## 4 Arizona West Mountain 2006 4.5 8
## 5 California West Pacific 2009 9.7 12
## 6 California West Pacific 2006 5 10
## 7 Colorado West Mountain 2009 6.1 2
## 8 Colorado West Mountain 2006 4.6 9
## 9 Hawaii West Pacific 2009 6.4 5
## 10 Hawaii West Pacific 2006 2.7 1
## # ℹ 16 more rowsWe have a bunch of columns here, with lots of repeated data. The first two rows show Alaska, and the state, region, and division columns are all identical. year is different, since there’s a row for 2006 and one for 2009, and unemployment and ranking are different, since those are the values for 2006 and 2009.
Now let’s pivot this wider so that we can subtract the ranking in 2006 from the ranking in 2009:
unemployment_west_rankings |>
pivot_wider(names_from = year, names_prefix = "rank_", values_from = ranking) |>
mutate(rank_diff = rank_2009 - rank_2006)
## # A tibble: 26 × 7
## state region division unemployment rank_2009 rank_2006 rank_diff
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Alaska West Pacific 7.1 7.5 NA NA
## 2 Alaska West Pacific 6.9 NA 13 NA
## 3 Arizona West Mountain 8.4 10 NA NA
## 4 Arizona West Mountain 4.5 NA 8 NA
## 5 California West Pacific 9.7 12 NA NA
## 6 California West Pacific 5 NA 10 NA
## 7 Colorado West Mountain 6.1 2 NA NA
## 8 Colorado West Mountain 4.6 NA 9 NA
## 9 Hawaii West Pacific 6.4 5 NA NA
## 10 Hawaii West Pacific 2.7 NA 1 NA
## # ℹ 16 more rowsThis didn’t work!
We still have two rows for each state. The first row has a value for rank_2009, but nothing for rank_2006; the second row has a value for rank_2006, but nothing for rank_2009. rank_diff is completely empty because you can’t calculate things like 7.5 - NA.
What happened here?
The reason this happened is that R doesn’t want to throw away or destroy any data. We told it to take the rank column and spread it wider so there’s a column for each year. Since state, region, and division are all the same for both years, it could collapse them into one row. But unemployment is different in each row/year, and it doesn’t know what to do with those values when pivoting, so it keeps them in two separate rows/years.
We can fix this two ways. First, we can get rid of any column that doesn’t repeat each year, like unemployment. That way R doesn’t have to worry about where to put those values in the wider data:
unemployment_west_rankings |>
select(-unemployment) |>
pivot_wider(names_from = year, names_prefix = "rank_", values_from = ranking) |>
mutate(rank_diff = rank_2009 - rank_2006)
## # A tibble: 13 × 6
## state region division rank_2009 rank_2006 rank_diff
## <chr> <chr> <chr> <dbl> <dbl> <dbl>
## 1 Alaska West Pacific 7.5 13 -5.5
## 2 Arizona West Mountain 10 8 2
## 3 California West Pacific 12 10 2
## 4 Colorado West Mountain 2 9 -7
## 5 Hawaii West Pacific 5 1 4
## 6 Idaho West Mountain 9 4 5
## 7 Montana West Mountain 6 5.5 0.5
## 8 Nevada West Mountain 11 5.5 5.5
## 9 New Mexico West Mountain 3.5 7 -3.5
## 10 Oregon West Pacific 13 12 1
## 11 Utah West Mountain 3.5 3 0.5
## 12 Washington West Pacific 7.5 11 -3.5
## 13 Wyoming West Mountain 1 2 -1Now there’s just one row for each state instead of two, since state, region, and division are identical across all the years.
The other way to fix it is to pivot both the ranking and the unemployment columns wider so that their values both have somewhere to go:
unemployment_west_rankings |>
pivot_wider(
names_from = year,
values_from = c(ranking, unemployment),
names_sep = "_"
) |>
mutate(rank_diff = ranking_2009 - ranking_2006)
## # A tibble: 13 × 8
## state region division ranking_2009 ranking_2006 unemployment_2009 unemployment_2006 rank_diff
## <chr> <chr> <chr> <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 Alaska West Pacific 7.5 13 7.1 6.9 -5.5
## 2 Arizona West Mountain 10 8 8.4 4.5 2
## 3 California West Pacific 12 10 9.7 5 2
## 4 Colorado West Mountain 2 9 6.1 4.6 -7
## 5 Hawaii West Pacific 5 1 6.4 2.7 4
## 6 Idaho West Mountain 9 4 7.2 3.8 5
## 7 Montana West Mountain 6 5.5 6.5 3.9 0.5
## 8 Nevada West Mountain 11 5.5 9.5 3.9 5.5
## 9 New Mexico West Mountain 3.5 7 6.2 4.4 -3.5
## 10 Oregon West Pacific 13 12 10.3 5.6 1
## 11 Utah West Mountain 3.5 3 6.2 3.5 0.5
## 12 Washington West Pacific 7.5 11 7.1 5.1 -3.5
## 13 Wyoming West Mountain 1 2 4.4 3.4 -1Now we have columns for ranking_2006 and ranking_2009 and unemployment_2006 and unemployment_2009, and we still have one row per state.
There were a couple common questions about seeds:
As discussed in the lecture, seeds make random things reproducible. They let you make random things again.
If you’ve ever played Minecraft, seeds are pretty important there too. Minecraft worlds (all the mountains, oceans, biomes, mines, etc.) are completely randomly generated. When you create a new world, it gives an option to specify a seed. If you don’t, the world will just be random. If you do, the world will still be random, but it’ll be the same random. There are actually Reddit forums where people play around with different seeds to find interesting random worlds—like weirdly shaped landmasses, interesting starting places, and so on. Some gamers will stream their games on YouTube or Twitch and will share their world’s seed so that others can play in the same auto-generated world. That doesn’t mean that others play with them—it means that others will have mountains and trees and oceans and mines and resources in exactly the same spot as them, since it’s the same randomly auto-generated world.
When R (or any computer program, really) generates random numbers, it uses an algorithm to simulate randomness. This algorithm always starts with an initial number, or seed. Typically it will use something like the current number of milliseconds since some date, so that every time you generate random numbers they’ll be different. Look at this, for instance:
They’re different both times.
That’s ordinarily totally fine, but if you care about reproducibility (like having a synthetic dataset with the same random values, or having jittered points in a plot be in the same position every time you render), it’s a good idea to set your own seed. This ensures that the random numbers you generate are the same every time you generate them.
If you set a seed, you control how the random algorithm starts. You’ll still generate random numbers, but they’ll be the same randomness every time, on anyone’s computer. Run this on your computer:
You’ll get 10, 6, and 5, just like I did here. They’re random, but they’re reproducibly random.
In data visualization, this is especially important for anything with randomness, like jittering points or repelling labels.
For instance, if we make this jittered strip plot of penguin data:
penguins <- palmerpenguins::penguins |> drop_na(sex)
ggplot(penguins, aes(x = species, y = body_mass_g)) +
# height = 0 makes it so points don't jitter up and down
geom_point(position = position_jitter(width = 0.25, height = 0))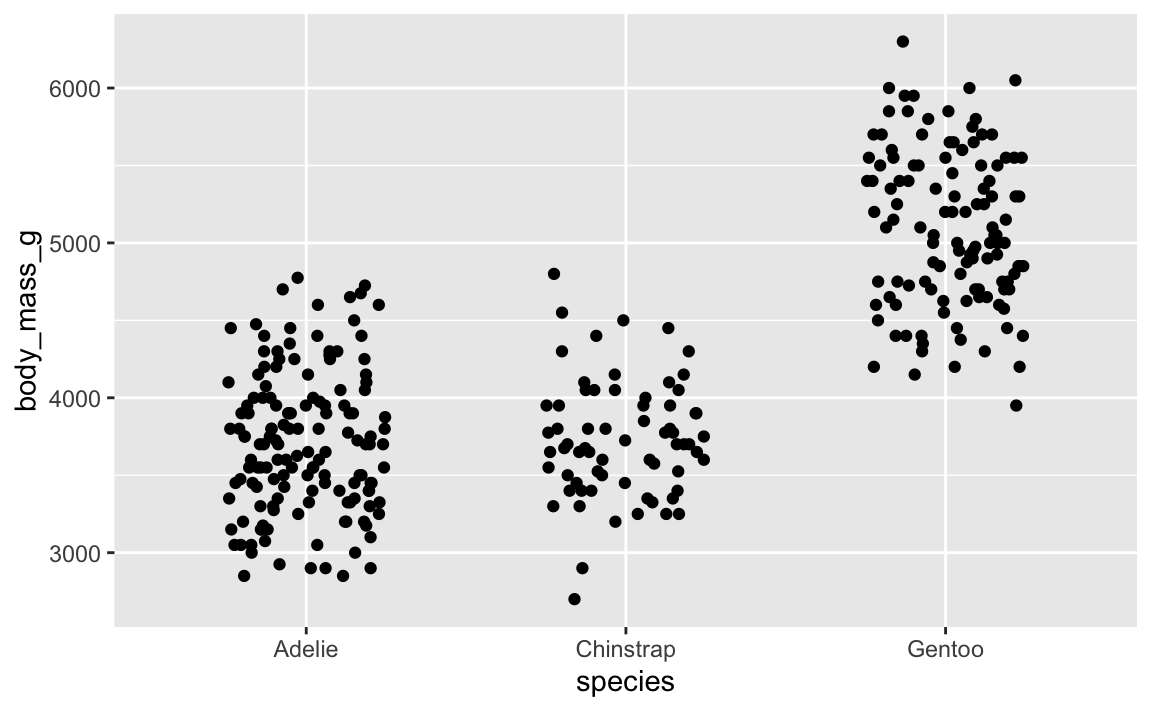
That looks cool. But if we rerender it:
ggplot(penguins, aes(x = species, y = body_mass_g)) +
geom_point(position = position_jitter(width = 0.25, height = 0))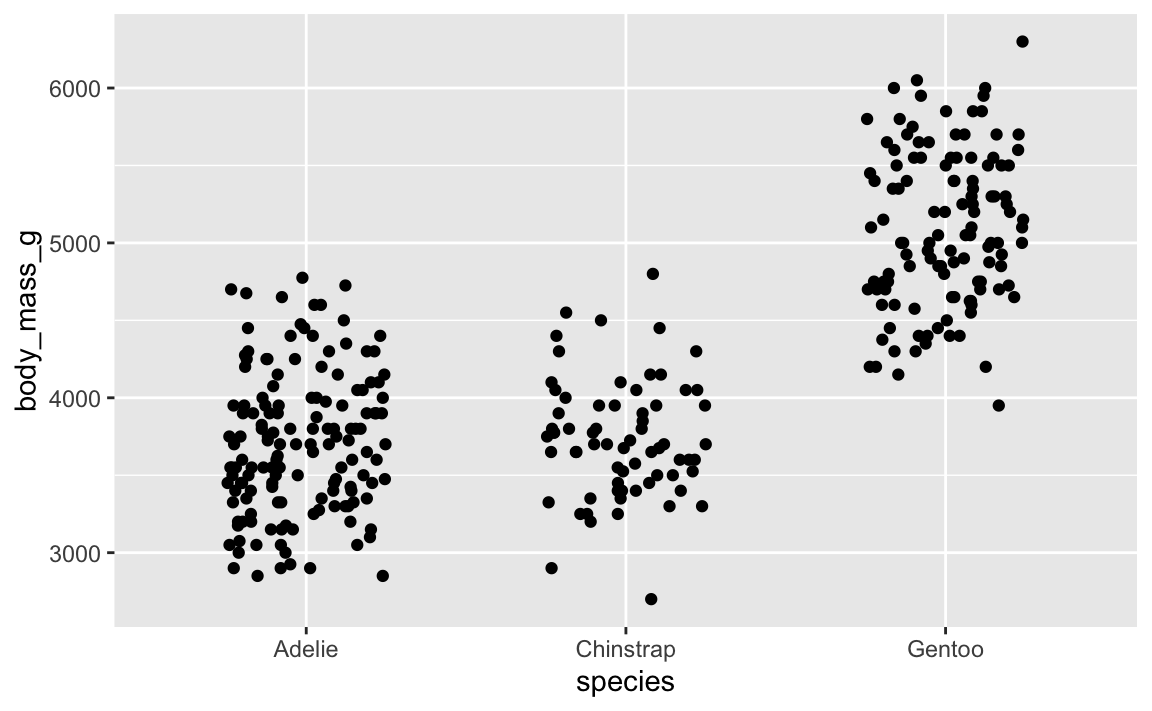
…it’s slightly different. And if we do it again:
ggplot(penguins, aes(x = species, y = body_mass_g)) +
geom_point(position = position_jitter(width = 0.25, height = 0))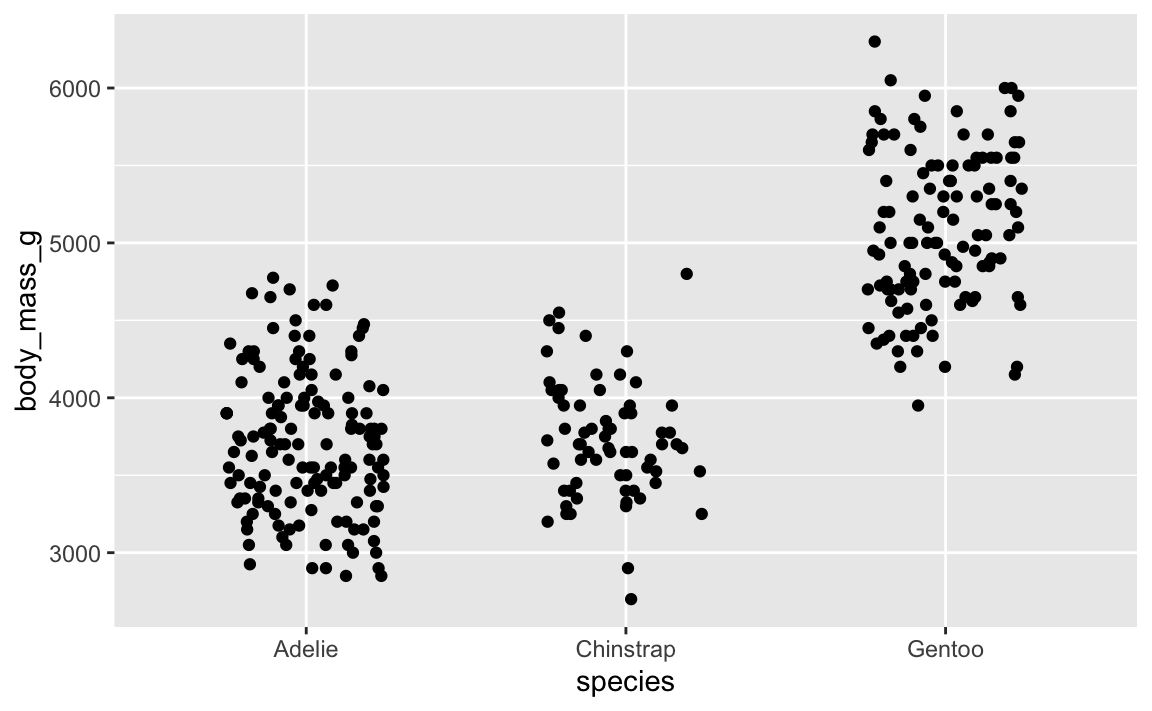
…it’s different again! It’s going to be different every time, which is annoying.
Slight variations in jittered points is a minor annoyance—a major annoyance is when you tinker with settings in geom_label_repel() to make sure everything is nice and not overlapping, and then when you render the plot again, everything is in a completely different spot. This happens because the repelling is random, just like jittering. You want the randomness, but you want the randomness to be the same every time.
To ensure that the randomness is the same each time, you can set a seed. position_jitter() and geom_label_repel() both have seed arguments that you can use. Here’s a randomly jittered plot:
ggplot(penguins, aes(x = species, y = body_mass_g)) +
geom_point(position = position_jitter(width = 0.25, height = 0, seed = 1234))
And here’s that same plot jittered again, but with the same randomness—it’s identical!
ggplot(penguins, aes(x = species, y = body_mass_g)) +
geom_point(position = position_jitter(width = 0.25, height = 0, seed = 1234))
You can also use set.seed() separately outside of the function, which sets the seed for all random things in the document (though note that this doesn’t create the same plot that position_jitter(seed = 1234) does! There are technical reasons for this, but you don’t need to worry about that.)
What should you use a seed? Whatever you want, really. In the slides, I had these as examples:
1234(567)113 (common (un)lucky number)42 (the answer to life, the universe, and everything)8675309 (Jenny’s number)24601 (Jean Valjean’s inmate number in Les Misérables)20250715 for something written on July 15, 2025In practice, especially for plots and making sure jittered and repelled things look good and consistent, it doesn’t really matter what you use.
Here’s what I typically do: